The programs engaged in this document were chosen as favorites (or in some cases, relics) of Collum group members. There is a large emphasis on those that are open source and free to use (listed in bold below), however there are a plethora of alternatives available across the internet and the following list should be taken as a starting point, not a superlative.
You’ll need the following programs for this course
Cornell VPN client (for Cornell students who intend to use the AS-CHEM cluster)
SFTP client (e.g. FileZilla, putty)
Molecular modeling program (e.g. GaussView, Avogadro, SAMSON, etc…)
Command-line terminal (e.g. the built in terminal on Linux/Mac OS or cygwin if you’re using a Windows machine)
Command-line text editor (e.g. Vim, Vi, Nano; helpful, but not required)
A text editor (e.g. Brackets, Sublime, VSCode, Notepad++, etc…)
3D-rendering/ray-tracing software, optional (e.g. CYLView, SAMSON, etc)
FileZilla
FileZilla is an SFTP (secure file transfer protocol) client that we’ll use to move files between our own computers and the cluster; we’ll have to do this since we can’t just wander over to the Baker server room and stick a USB drive into the stacks every time we want our data.
Setup
If you haven’t talked to your system admin to set yourself up a cluster account then stop here and get that done first. You’ll need a fully set up account (on our cluster or your own) to get the most out of this course.
These setup instructions are for the AS-CHEM cluster only, if you’re not trying to connect to this cluster, contact your system administrator for instructions on how to transfer files to/from your own system.
- Install and connect to the Cornell VPN
- Download the latest FileZilla release. Follow these instructions to install it.
- Start FileZilla and open the site manager by going to File > Site Manager.
- Add a new connection by clicking “New Site”

- Configure the settings for the new site as shown:
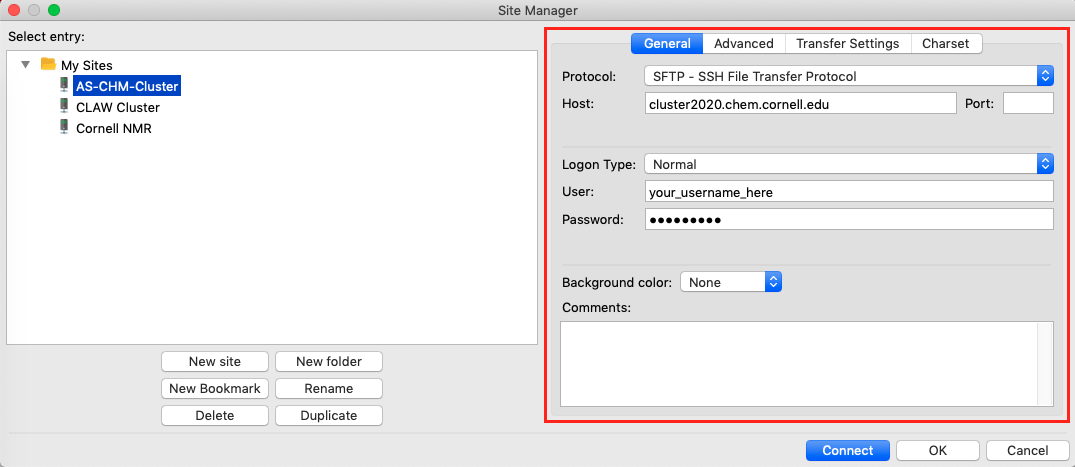
- Protocol:
SFTP - SSH File Transfer Protocol - Host:
cluster2020.chem.cornell.edu - Logon type:
NormalorAsk for passwordif you want to be asked every time - User: Your cluster logon (typically your netID)
- Password: If you selected “Normal” for logon type then enter your cluster password. If “Ask for password” then you will be prompted for your password every time you connect.
- Protocol:
- Click
Connectto connect to the cluster and make sure you have everything set up correctly. Your cluster home directory should show up on the right.
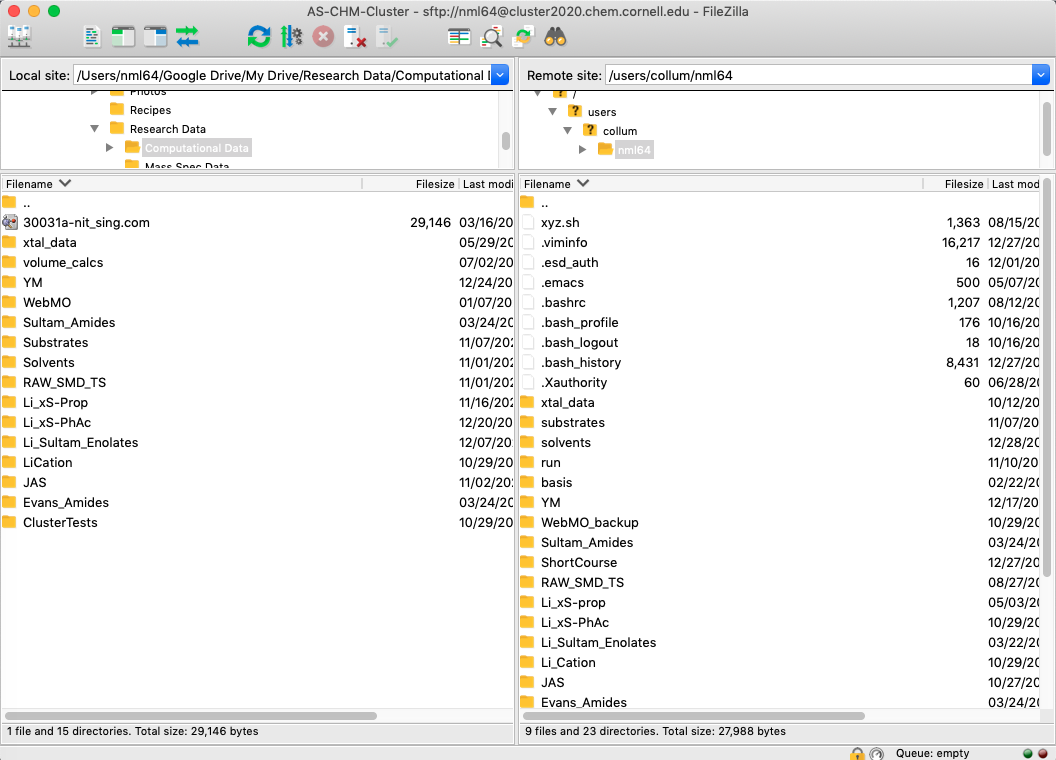
To reconnect in the future just go back to the site manager, select the cluster site and click Connect. Remember that you need to be connected to the Cornell VPN to access the cluster.
Command-line terminals
If you are working on a Mac or Linux machine feel free to skip this part. Linux distributions and MacOS come preinstalled with a command-line terminal.
Technically, Windows machines come with PowerShell (which is its own shell and scripting language (two things you’ll learn about in the next lesson)), but for the sake of uniformity and since the cluster runs on CentOS, we’ll use a bash shell. Windows users should download and install cygwin. By default cygwin does not come with text editors installed (don’t ask me why) so you’ll have to go through the setup.exe program to install these packages (start with vim and nano). This blog post may come in handy.
Molecular modeling programs
GaussView
Due to licensing constraints you’ll need to visit ChemIT to get GaussView installed on your computer.
Avogardo
Avogadro is a free, open-source molecular editor and visualizer that can be used as an alternative to GaussView. It comes fully documented and has built-in compatibility with Gaussian input adn output. Download and install it from SourceForge.
Text Editors
Brackets
Brackets is an open-source, lightweight text editor that was built for web designers and front-end developers, but it has access to a the local filesystem a really helpful feature in keeping our files organized. Check out the Brackets Wiki to learn more (always start with the README).
VSCode
Visual Studio Code is what’s known as an IDE, for integrated development environment. It is open-source, but not lightweight. VS Code is super powerful, with built-in features like line-by-line debugging, intelligent code completion, built-in terminal, expanded language support, and spell check. If you’re just getting started, this is not the text editor you’re looking for, but as you become more experienced you may find that you want more than a simple code editor. Read the docs to get started.
3D-rendering and ray-tracing
CYLView
CYLView is a free molecular visualizer currently in development by Claud Y. Legault at the University of Sherbrook (Canada). Invaluable for drawing publication-quality chemical structures from computational output. Both versions CYLView1.0 and CYLView20 can be downloaded here. Mac users will need to install XQuartz in order to use CYLView1.0. Its recommended that you install both versions as CYLView20 is still pretty barebones.
Linux Basics |